

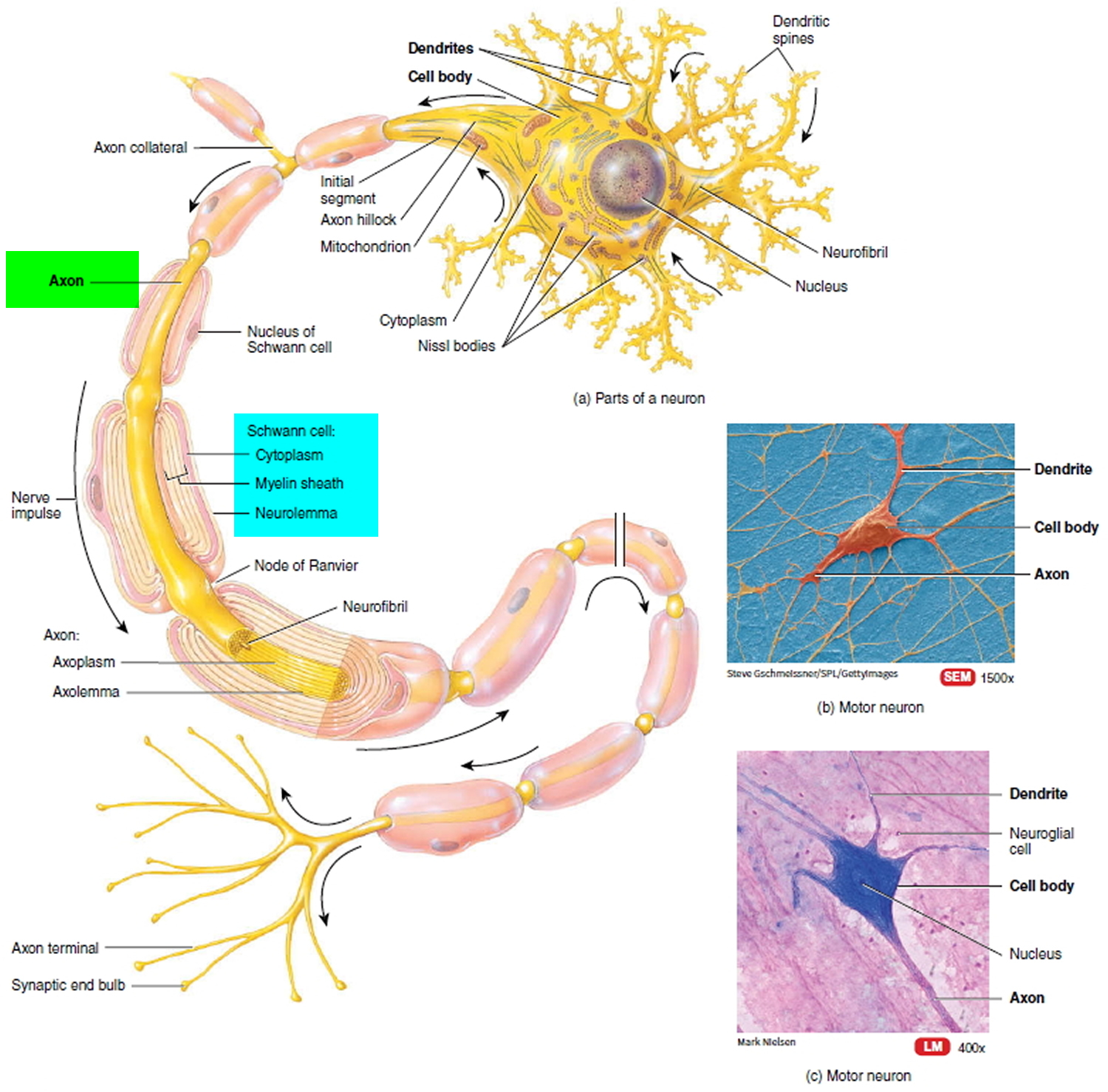
One approach to augment EM datasets with functional information is to physiologically record from neurons in a tissue sample prior to fixation ( Bock et al., 2011 Briggman et al., 2011 Lee et al., 2016 Wanner and Friedrich, 2020). Connectivity data derived from EM, however, does not alone contain sufficient information about the functional properties of neurons to fully constrain biologically plausible computational models. Recent improvements in automating the collection of 3D electron microscopy (EM) data have dramatically increased the tissue volumes that can be acquired ( Briggman and Bock, 2012 Eberle et al., 2015).
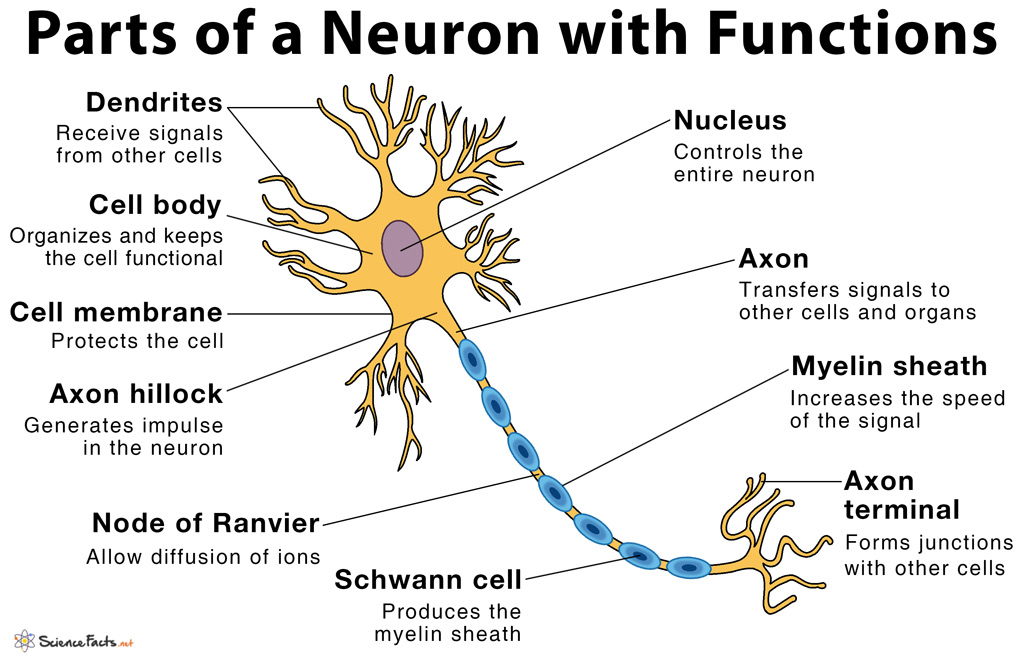
Anatomical reconstructions of synaptic connectivity are one essential component of understanding neuronal circuits in the nervous system.


 0 kommentar(er)
0 kommentar(er)
